deltaDOT TECHNIQUES : Protein fingerprinting
Rationale: In each of the above cases the interpretation of a complex sample with many (e.g. 40+) proteins needs to have more controlled measurements than is commonly available with canonical techniques. This leads to many issues, not least that of the difficulty of standardisation across the industry due to the lack of non-subjective techniques. An example of this might be demonstrated by the need to monitor the expression of an engineered protein in CHO cells. At what point does the DNA construct and its associated regulatory factors yield a statistically significant increase in the target protein? What other proteins are affected by the introduction of the construct and how does that affect the overall cell expression system? These parameters need a more standardised data interpretation than an experienced (but ultimately subjective) ‘eye’ can provide in order to allow the development of standard operational procedures in biotherapeutics production.
deltaDOT Solution: Capillary Gel Electrophoresis (CGE) using Label Free Intrinsic Imaging (LFII® ) with statistical analysis supplies a hard number objective metric for protein profiling. CGE is a quick, cost-effective method for characterising complex protein samples and assessing the effect of process modifications on the final product. This technique is also used to monitor consistency between batches, yield optimisation, purity analysis and for stability studies. Traditionally, slab gels have been used to assess variations in protein profiles, but these are generally labour-intensive and expensive due to the complex sample preparation steps required and costly reagents and consumables. CGE data produced on the deltaDOT HPCE-512 capillary electrophoresis instrument comparing different protein products is presented (Figure 1). Our strategy for assessing differences in complex samples involves identifying the most abundant peaks within the sample for quantification. The regions between these peaks are also quantified so that any changes in the less abundant proteins can be detected (Figure 1A). Repeatability and intermediate precision were assessed by carrying out 5 consecutive injections of the sample per day on two separate days (Figure 1B). Excellent reproducibility and intermediate precision were observed with the samples, with relative standard deviation (RSD) values between 1% and 5% across both days. Differences in profiles were assessed by comparing percentage peak areas of the main peaks. Clear differences were observed between the samples (Table 1). Different methods for normalisation of the data were tested, including the use of % area, % RSD values were then calculated (Table 2).
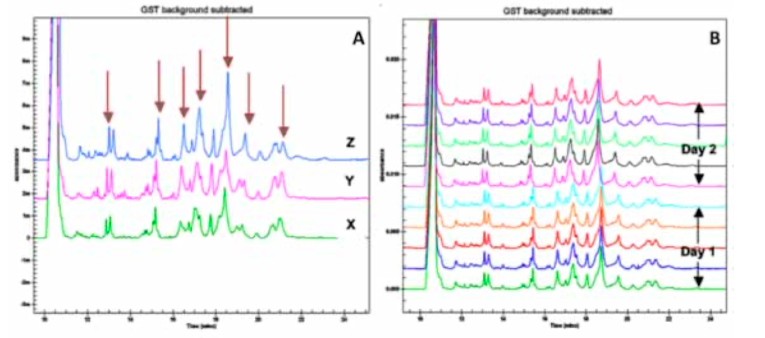
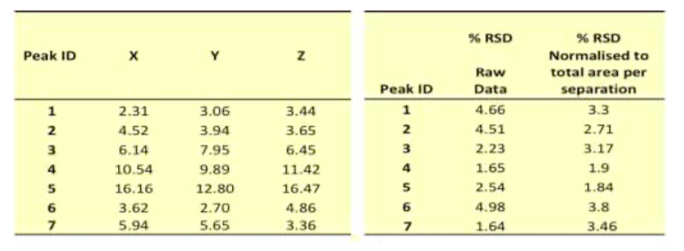
Following assessment of the repeatability and precision of the instrument, acceptance criteria can be assigned to different batches of the product to ensure they fall within tolerance. These tolerance limits can be set take into account the statistical likelihood that observed deviations are due to the statistical variations between experiments (the precision of the instrument) and the allowable differences between batches as set by the manufacturer.
Commercial applications: This application has been applied in many areas, as follows.
• Batch release comparison analysis of a complex heat-shock protein vaccine formulation.
• Analysis of stability between innovator and originator mAb/biosimilar samples.
• Expression construct optimisation in synthetic biology
• Comparison of bioreactor growth parameters for stem cell production, allowing a statistical metric to be assigned for each method allowing robust evaluation.
• Characterisation of viral gene therapy vaccine vectors. Having assessed viral titre by one form of CE (CZE) the virus is disrupted and analysed by CGE to establish differences in the protein profiles of the samples.
For any additional information on our ability to characterise complex samples please visit our website (www.deltadot.com) or contact us on info@deltadot.com.
